Menu
Microsatellites (MS) are repetitive nucleotide sequences distributed at millions of loci across our genome1. Because of their repetitive sequence, they are prone to DNA polymerase slippage events during DNA replication, resulting in repeat length variations1. Proteins in the highly conserved mismatch repair (MMR) pathway usually identify and repair these types of errors, safeguarding the genome from potentially deleterious mutations2. If this repair machinery fails, for example, due to genetic or epigenetic loss of MMR protein function, the rate of spontaneous mutations on MS regions throughout the genome increases1. Therefore, microsatellite instability (MSI) is considered a molecular fingerprint for defects in the mismatch repair system (dMMR) (Figure 1) and is associated with various malignancies1.

A recent study on more than 11,000 tissue samples covering 39 cancer types showed that microsatellite instability (MSI) was present within 27 cancer types (3.8% of all cancers analyzed). Twelve cancer types were found with an MSI prevalence more significant than 1%, represented mainly by endometrial (31.4%), gastric (19.1%), and colorectal (CRC) adenocarcinomas (16.0%)3. Most MSI tumors arise sporadically4, and other cases result from inherited cancer predisposition syndromes such as Lynch syndrome (LS)5.
MSI has several implications in the management of patients with solid cancers4. First, detection of MSI can be used as a screening strategy for LS, as almost 95% of the LS-associated malignancies have MSI 5. Secondly, MSI status represents a positive prognostic value for localized CRC and gastric cancers6. Finally, in recent years, MSI has gained considerable attention as a predictive marker of immune checkpoint inhibitor therapy response across multiple tumor types5,6.
Identification of MSI relies on comparing the size of selected microsatellites in normal and tumor tissue of the same individual. The extent of the repeat length variation determines the MSI level of the assessed tumor, which can then be classified as MS-instable or MS-stable6.
Despite its recognized pan-cancer value, commonly used methods only support MSI detection in colorectal cancers, since their sensitivity in other tumor types is low. In addition to the tissue-specific differences that impact the sensitivity of MSI detection, the analytical performance is also impacted by patient ethnicity, tumor content, and other sample-specific properties3.
Immunohistochemistry (IHC) and PCR-based assays performed on tumor tissue samples represent the two current reference methods for MSI/dMMR status detection. Being MSI strictly correlated with the proper functioning of the MMR pathway, the immunohistochemistry (IHC) detection of MMR proteins allows to assess MSI and thus determine MS stable or unstable cases1. Low cost and wide availability are the main advantages of this method. However, this test cannot be combined with other molecular diagnostics tests and is also known to provide false positive and negative results due to different technical and biological factors3, 8.
A complementary and rather preferred tool to investigate MSI is polymerase chain reaction (PCR) analysis followed by capillary electrophoresis analysis, which amplifies informative selected MS and then identifies fragment length polymorphisms2. For the PCR to be indicative, standardized panels of highly unstable mononucleotide or dinucleotide repeats should be used. In the case of colorectal cancers, the Bethesda/ National Cancer Institute (NCI) panel of five repeats has been considered the gold standard for many years9. However, because of the extensive tumor specificity of MSI targets, the use of the same small subset of markers becomes inadequate for other cancer types1.
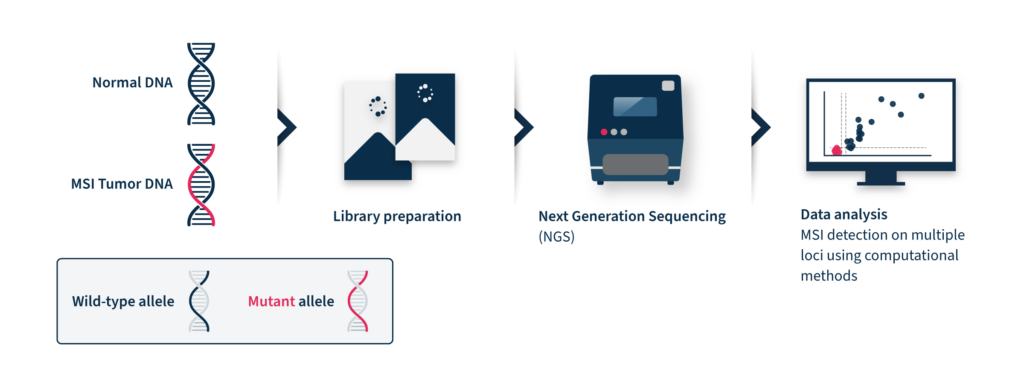
Next-generation sequencing (NGS)-based MSI detection methods allow simultaneous analysis of a larger number of microsatellite regions for each patient with a higher sensitivity for low prevalence mutations2. In addition, NGS enables the analysis of other cancer-related molecular signatures and genetic lesions with one unique assay, facilitating the adoption of MSI clinical testing and supporting personalized cancer therapy8. Despite these advantages, the analysis of MSI through NGS is challenging. NGS accuracy can be low by analyzing tumor-only data due to the inter-and intra-tumor differences in the frequency and position of MSI diagnostic events2. These differences limit the analytical performance of this method, which relies on a baseline reference distribution to determine MSI samples. Several computational approaches have been developed until now to analyze MSI data coming from NGS, considering the difficulties of microsatellite sequences, including management of stutter peak formation induced by PCR amplification during the library preparation step, sequencing errors induced by homopolymers, and the shortcomings of sequencing read length that limit the length of the microsatellites analyzed (Figure 2).
Therefore, NGS-based MS assessment requires expensive and time-consuming bioinformatic analysis with proper algorithms for data interpretation9.
At SOPHiA GENETICS, we believe that data-driven medicine has the potential to improve health outcomes. That’s why we developed MUSTARD™, an algorithm with comparable sensitivity to PCR-based methods and only requiring tumor data to function. MUSTARD™ is a curve-fitting algorithm that aims at better identifying differences in the read length distribution of MSI and normal samples.
MUSTARD™ was effectively developed to perform MSI classification in multiple cancers and trained on a specific selection of more than 100 microsatellite regions identified as highly performant in different cancer types. This means that, across several tumors, the length of these loci significantly differed from MS stable samples. Using such a subset of loci as a reference, the analytical performance of MUSTARD™ increased and became comparable to that of other NGS-based solutions with the great advantage of requiring tumor data only for the analysis.
MUSTARD™ is already available in the SOPHiA DDM™ for TSO500 bioinformatic workflow.
SOPHiA DDM™ has been indeed optimized to detect SNVs, Indels, CNVs, and gene fusions simultaneously to evaluate MSI and tumor mutational burden (TMB), covered by the Illumina TruSight® Oncology 500 (TSO500). The MSI algorithm has been extensively tested on 74 samples, with known MSI status from at least 9 cancer types and 7 different centers. The internal analytical validation study showed 100% sensitivity and specificity, reproducing the MMR/MSI status detected by IHC. Moreover, in a comparison study, MUSTARD™ outperformed the current standard analytical technology for TSO500. In contrast to the standard analytical technology score, which does not allow clear separation between samples classified by IHC as dMMR and pMMR (horizontal line), MUSTARD™ score distribution for these two populations is distinct (vertical lines), (Figure 3).
MUSTARD™ has proven to overcome the main challenges currently faced by most NGS-based solutions and make accurate and extensive MSI detection more attainable than ever. Read more about our MSI detection approach in our Frontiers in Oncology publication.

SOPHiA GENETICS products are for Research Use Only and not for use in diagnostic procedures unless specified otherwise.
SOPHiA DDM™ Dx Hereditary Cancer Solution, SOPHiA DDM™ Dx RNAtarget Oncology Solution and SOPHiA DDM™ Dx Homologous Recombination Deficiency Solution are available as CE-IVD products for In Vitro Diagnostic Use in the European Economic Area (EEA), the United Kingdom and Switzerland. SOPHiA DDM™ Dx Myeloid Solution and SOPHiA DDM™ Dx Solid Tumor Solution are available as CE-IVD products for In Vitro Diagnostic Use in the EEA, the United Kingdom, Switzerland, and Israel. Information about products that may or may not be available in different countries and if applicable, may or may not have received approval or market clearance by a governmental regulatory body for different indications for use. Please contact us at [email protected] to obtain the appropriate product information for your country of residence.
All third-party trademarks listed by SOPHiA GENETICS remain the property of their respective owners. Unless specifically identified as such, SOPHiA GENETICS’ use of third-party trademarks does not indicate any relationship, sponsorship, or endorsement between SOPHiA GENETICS and the owners of these trademarks. Any references by SOPHiA GENETICS to third-party trademarks is to identify the corresponding third-party goods and/or services and shall be considered nominative fair use under the trademark law.