Lymphoid neoplasms encompassing lymphomas and some leukemia like Chronic Lymphocytic Leukaemia (CLL) are the most common type of blood cancer 1.
With increasing evidence for the stratification of tumor types with distinct clinical and biological features according to biomarkers, and the progress in targeted therapy, tailored NGS-based workflows empower experts to get high-quality and reproducible data to accelerate their studies.
Expertly designed gene panel, customizable to meet your laboratory’s specific research needs
Detection of challenging variants including SNVs, Indels, CNVs and Immunoglobulin (IGH) rearrangements
Tailored probes to maximize on-target rate and coverage uniformity, for a cost-effective multiplexing and sequencing
Ready-to-use target-enriched library in just 2 days
Data analysis from FASTQ files in as little as 4 hours
Streamlined workflow, from sample to report, to accelerate your analysis
Easy variant visualization, classification and annotation within the SOPHiA DDM™ Platform
Tertiary analysis based on the latest scientific evidence on the relevant variants with OncoPortal™Plus
Access to SOPHiA GENETICS Community, one of the largest networks of connected healthcare institutions, to gain and share scientific knowledge
Accurately characterize lymphoid malignancies
Lymphoid neoplasms originate from hematopoietic disruptions in the lymphoid lineage due to complex mutations. Improvements in sequencing technics and the development of targeted therapies call for a refined classification according to genetic alterations2. Therefore, advances in cancer study depend on timely, cost-effective, and reliable high-throughput sequencing strategies.
Lymphomas
Acute lymphocytic leukemia (ALL)
Chromosomal rearrangements such as the BCR::ABL1 (Philadelphia chromosome ) or ETV6::RUNX1 are hallmarks of ALL. Recurrent fusions involving kinase genes such as ABL1, ABL2, PDGFRB, CSF1R, CRLF2, JAK2, and EPOR are also observed3.
Chronic lymphocytic leukemia (CLL)

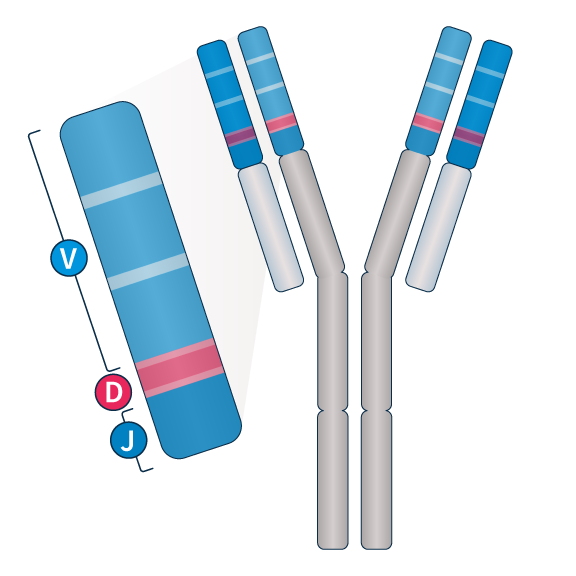
Get the whole Immunoglobulin picture
Mature B cells express a unique immunoglobulin gene heavy chain (IGH) rearrangement of the V(D)J region. The clonotype as well as the mutational status of IGH are critical biomarkers for tumor stratification. Our optimized probes capture this challenging region in one assay and allow for further characterizations following international recommendations5.
Confidently assess challenging biomarkers
Molecular profiling by NGS has introduced a paradigm shift in investigating the pathogenic variants causing different lymphoid malignancies. Indeed, it allows the characterization of multiple SNVs, Indels, and CNVs in one unique experiment. With an optimized probe design and the advanced analytical capabilities of the SOPHiA DDM™️ Platform, challenging variants in GC-rich and complex regions such as EGR2, NFKBI and IGH locus are covered.
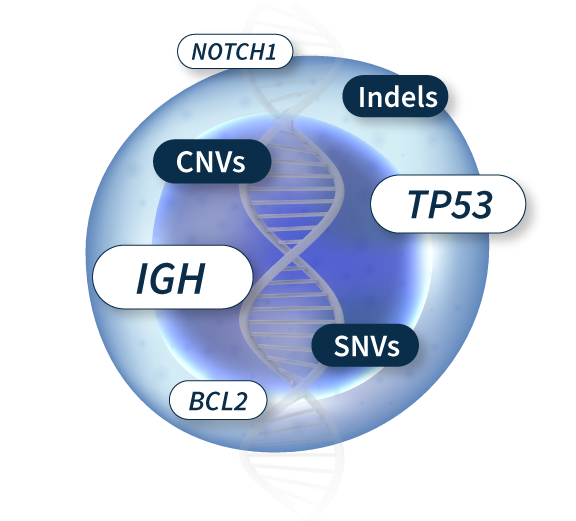

Trust your data
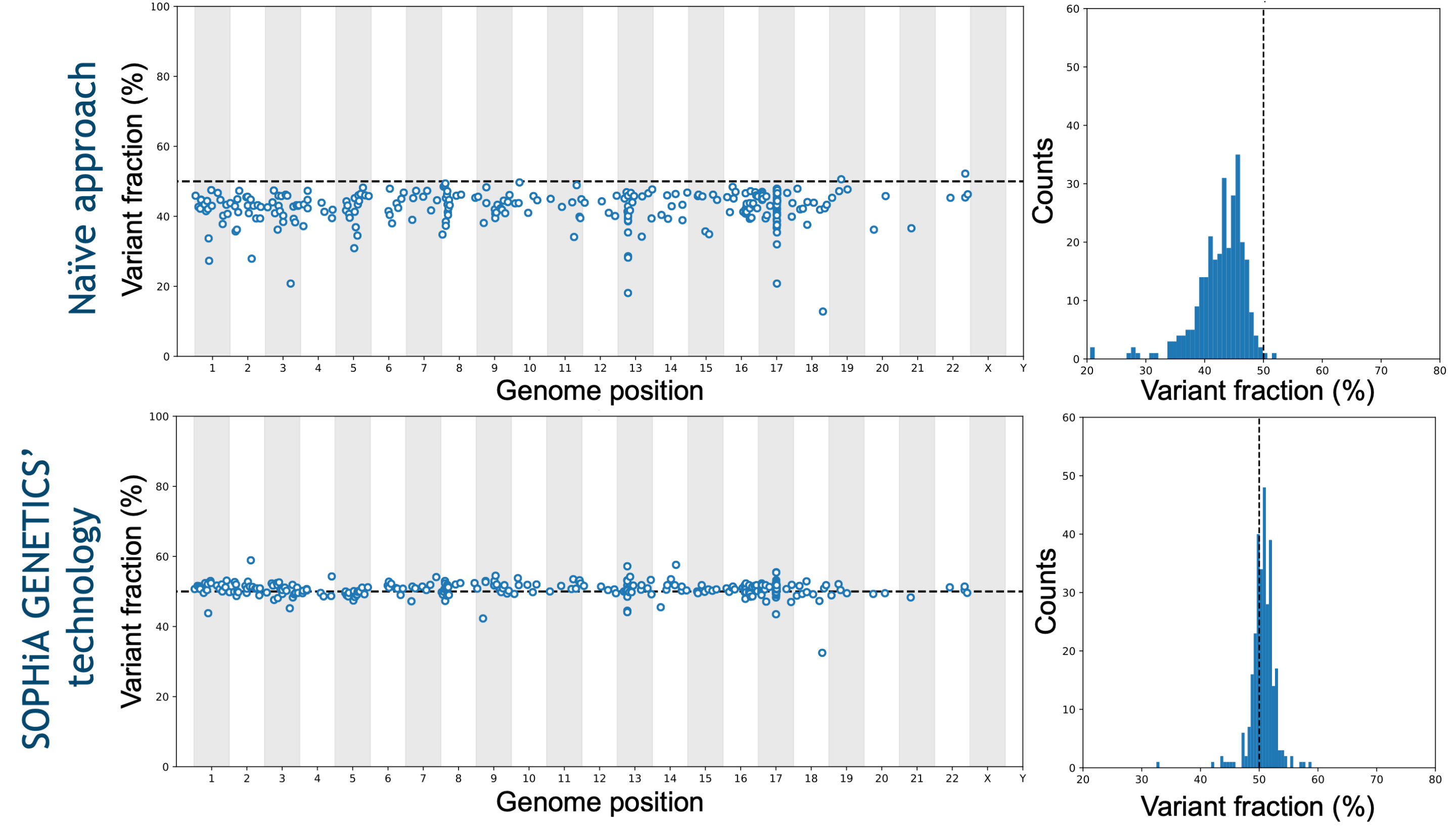
Exceptional analytical performance
Addressing the technical limitations related to the analyses of key biomarkers, the SOPHiA DDM™️ solutions combined with the SOPHiA DDM™️ Platform exhibit advanced analytical capabilities.
Data on file.
Trust your data
Optimum probe design
Because good probes do not make a perfect assay, SOPHiA GENETICS’ technology takes into account genomic variability in probe design to reduce coverage bias and optimize binding.
Data on file.
Trust your data
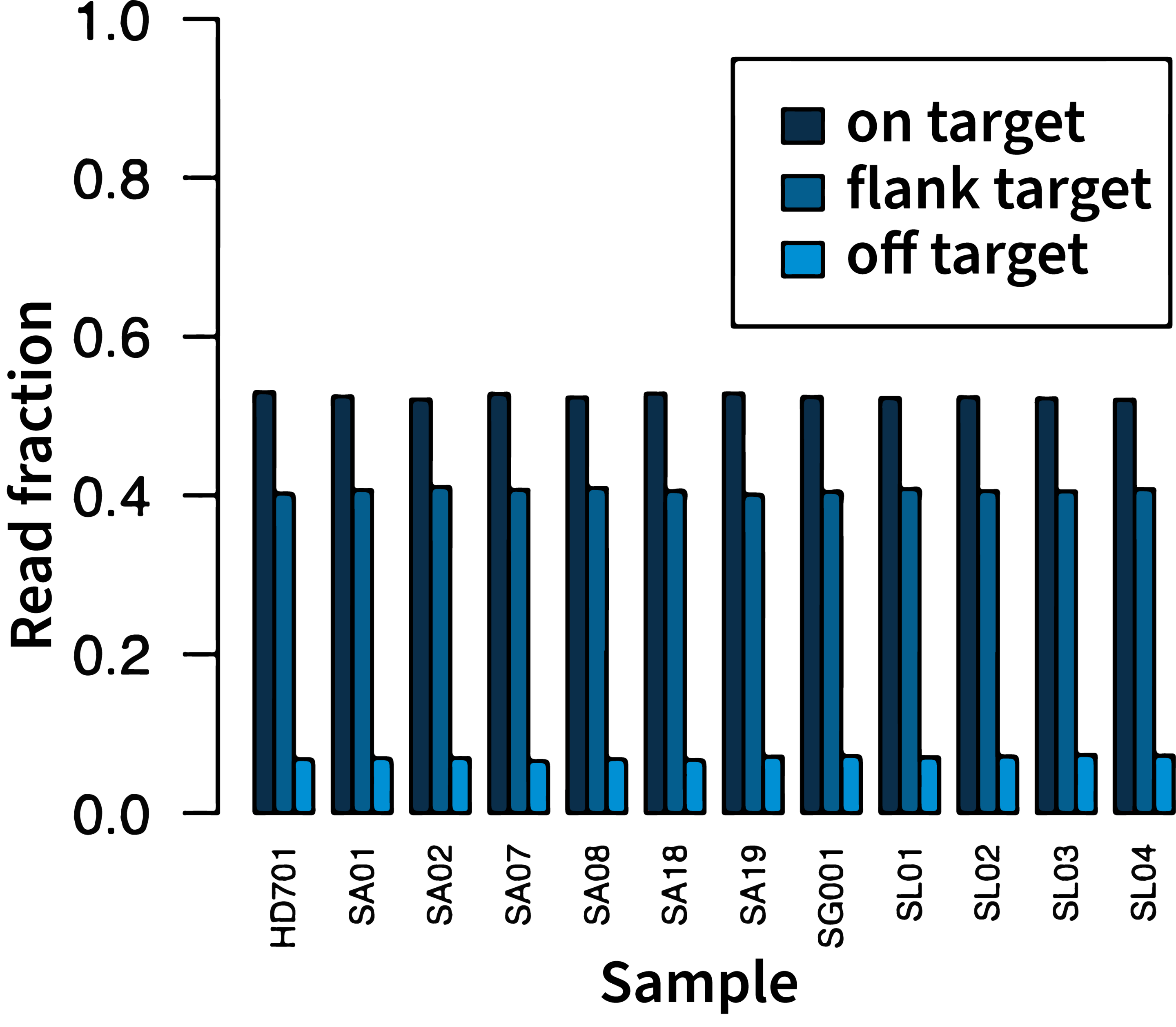
High on-target rate
In a typical run, the on-target rate (on-target + flanking target reads) is over 87%, maximizing the number of useful reads and increasing the multiplexing capacity.
Data on file.
Trust your data
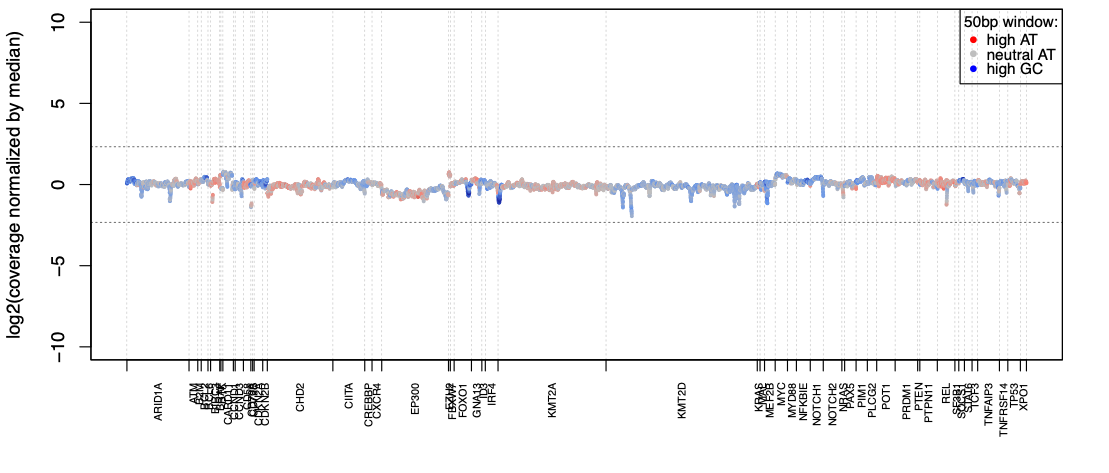
Excellent coverage uniformity
SOPHiA DDM™️ applications reach over 99% coverage uniformity across AT- and GC-rich regions of the targeted genes, making each sequencing extremely cost-efficient.
Data on file.
Trust your data
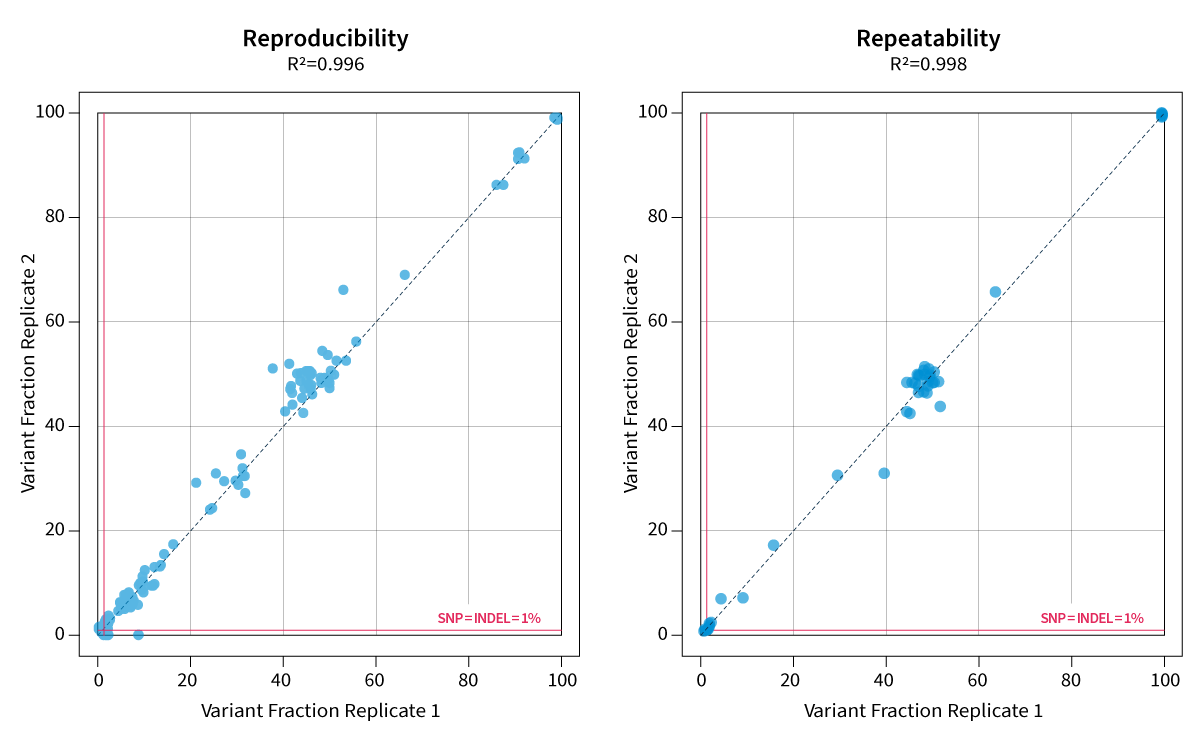
High reproducibility and repeatability
With almost 100% reproducibility and repeatability, SOPHiA DDM™️ Community CLL Clonality Solution provides consistent inter- and intra-runs results, insuring stable and trustworthy sequencing data.
Data on file.
Choose the solution that fits your needs
Our portfolio offers a range of ready-to-use yet customizable solutions that enable precise characterization of lymphoid malignancies driver genes by targeting key variants from FFPE, blood or bone marrow samples.
SOPHiA DDM™️ Lymphoma Solution
SOPHiA DDM™️ Lymphoma Solution is a comprehensive application that covers 54 genes associated with B- and T-cell lymphomas such as diffuse large B-cell, follicular, mantle cell and Burkitt lymphoma.
SOPHiA DDM™️ Community CLL Clonality Solution
SOPHiA DDM™️ Community CLL Clonality Solution is an expertly designed panel to specifically target 23 relevant genes for CLL, including the guidelines recommended biomarkers TP53 mutational status and IGH rearrangements.
SOPHiA GENETICS™ Community Panels for Blood Cancers
SOPHiA GENETICS™ Community Panels for Blood Cancers are targeted panels developed and tested by genomic experts in hematological malignancies to minimize set-up challenges and accelerate your workflow.
It is a great satisfaction to see how thanks to the partnership with SOPHiA GENETICS and Diagnóstica Longwood we will be able to transfer into clinical practice the relevant information generated for so many years in the research of CLL using a simple and robust assay.

Elias Campo, MD, PhD
Principal investigator and director of IDIBAPS and FCRB, Spain
Specifications
| Parameters | SOPHiA DDM™ Lymphoma Solution (LYS) | SOPHiA DDM™ Community CLL Clonality Solution |
|---|---|---|
| Associated disorders | B- and T-cell lymphomas such as diffuse large B-cell, follicular, mantle cell, and Burkitt lymphomas | Chronic Lymphocyte Leukaemia |
| Genes | 54 genes: ARID1A, ATM, B2M, BCL2, BCL6, BIRC, BRAF, BTK, CARD11, CCND1, CCND3, CD58, CD79A, CD79B, CDKN2A, CDKN2B, CHD2, CIITA, CREBBP, CXCR4, EP300, EZH2, FBXW7, FOXO1, GNA13, ID3, IRF4, KMT2A, KMT2D, KRAS, MAL, MEF2B, MYC, MYD88, NFKBIE, NOTCH1, NOTCH2, NRAS), PAX5, PIM1, PLCG2, POT1, PRDM1, PTEN, PTPN11, REL, SF3B1, SOCS1, STAT6, TCF3, TNFAIP3, TNFRSF14, TP53, XPO1 | 23 genes: ATF1, ATM, BCL2, BIRC3, BTK, CDK4, CUL4A, CXCR4, DLEU1, EGR2, FBXW7, KLF5, KRAS, MYD88, NFKBIE, NOTCH1, PLCG2, POT1, PROZ, RB1, SF3B1, TP53, XPO1 + IGH rearrangement |
| Target Region Size | 118 Kb | 48 Kb |
| Key Biomarkers | CREBBP, EP300, EZH2, MEF2B, MYD88, MYC, BRAF | IGH rearrangement, TP53, BTK, BCL2, NOTCH1 |
| Sample Type | FFPE, blood, and bone marrow | Blood and bone marrow |
| Starting material | 50 ng DNA | 200 ng DNA |
| Sequencer Compatibility |
Illumina MiSeq® and NextSeq® |
Illumina MiSeq®, NextSeq® and MiniSeq™️ |
| Library Preparation Time | 1.5 days | 1.5 days |
| Analysis Time From FASTQ | 4 hours | 4 hours |
| Detected Variants |
|
|
Resources
Fact sheets and flyers
Want to know more?
Get in touch with us.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. http://dx.doi.org/10.3322/caac.21660
- Alaggio R, Amador C, Anagnostopoulos I, Attygalle AD, Araujo IB de O, Berti E, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid neoplasms. Leukemia. 2022;36(7):1720–48. http://dx.doi.org/10.1038/s41375-022-01620-2
- Moorman AV. New and emerging prognostic and predictive genetic biomarkers in B-cell precursor acute lymphoblastic leukemia. Haematologica. 2016;101(4):407–16. http://dx.doi.org/10.3324/haematol.2015.141101
- Eichhorst B, Robak T, Montserrat E, Ghia P, Niemann CU, Kater AP, et al. Chronic lymphocytic leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2021;32(1):23–33. http://dx.doi.org/10.1016/j.annonc.2020.09.019
- Agathangelidis A, Chatzidimitriou A, Chatzikonstantinou T, Tresoldi C, Davis Z, Giudicelli V, et al. Immunoglobulin gene sequence analysis in chronic lymphocytic leukemia: the 2022 update of the recommendations by ERIC, the European Research Initiative on CLL. Leukemia. 2022;36(8):1961–8. http://dx.doi.org/10.1038/s41375-022-01604-2
- Bosch F, Dalla-Favera R. Chronic lymphocytic leukaemia: from genetics to treatment. Nat Rev Clin Oncol. 2019;16(11):684–701. http://dx.doi.org/10.1038/s41571-019-0239-8